The protocols, using a minimum of 100 pg and creating fragment sizes 100 bp 1kb. Greninger AL, Naccache SN, Federman S, Yu G, Mbala P, Bres V, Chiu CY. Template preparation is unique in the PacBio process as it involves production of a SMRTbell, a circular double-stranded DNA molecule with a known adapter sequence complementary to the primers used to initiate the DNA synthesis on the template. Greenough L, Schermerhorn KM, Mazzola L, Bybee J, Rivizzigno D, Cantin E, Gardner AF. In general, most of these new SBS methods do not use dideoxy terminators, although reversible terminators are used in some technologies, which allow the nucleotide incorporation reactions to proceed normally while imaging the incorporated nucleotides, and then removing synthesis blocking moieties on the labeled nucleotides to allow the incorporation of the next base in the sequence. The intensity of light also provides information concerning homopolymer runs of nucleotides in the sequence, although difficulties are encountered with larger tracts of the same nucleotide. Drmanac R, Drmanac S, Chui G, Diaz R, Hou A, Jin H, Little D. Sequencing by hybridization (SBH): advantages, achievements, and opportunities. Ordered restriction maps of Saccharomyces cerevisiae chromosomes constructed by optical mapping. Thus runs of a single nucleotide can also be determined. Sequencing by synthesis (SBS) methods have taken several distinct approaches (Fuller et al., 2009; Mardis, 2008; Metzker, 2010; Quail et al., 2012; Shendure & Ji, 2008). Cao MD, Ganesamoorthy D, Elliott AG, Zhang H, Cooper MA, Coin LJ. In order to control the device, a laptop with pre-loaded software is provided. When the complex encounters a nanopore, one DNA strand enters the nanopore and the translocation rate through the pore is regulated by DNA polymerase synthesis and translocation. Several versions of their machines are available, the Pippin Prep, BluePippin, or PippinHT, in which fragments of selected lengths are collected, eliminating all others. 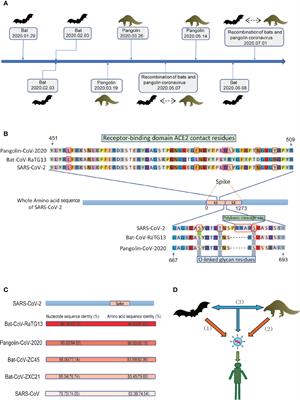 The second generation methods generally use a solid support containing micro channels or wells in which the sequencing reactions occur. PMC legacy view The next nucleotide can then be incorporated. The reagent costs are usually dependent on the volume ordered and are often negotiated with the vendor. In terms of throughput, the PacBio RS II (2013) uses chips with 150,000 ZMWs, which when optimized, can generate as much as 350 megabases of sequence per SMRT cell. Sequencing by electron microscopy has not yet been commercialized. The methylomes of six bacteria. Often, PacBio assemblies are combined with other methods, such as Illumina sequencing, for increased accuracy. Higher order multiplexing (barcoding) will enable more samples to be processed in a shorter time and at reduced cost. Mostovoy Y, Levy-Sakin M, Lam J, Lam ET, Hastie AR, Marks P, Kwok PY. In theory, more than a hundred kb of DNA could be threaded through the nanopore, and with many channels, tens to hundreds of Gb of sequence could be achieved at relatively low cost. The challenges of sequencing by synthesis. It is possible to pass long DNA molecules through small diameter holes and measure differing currents as each nucleotide passes by a linked detector (Benner et al., 2007; Branton et al., 2008; Cherf et al., 2012; Hornblower et al., 2007; Kasianowicz, Brandin, Branton, & Deamer, 1996; Liu, Wang, Deng, & Chen, 2016); see (https://www.youtube.com/watch?v=GUb1TZvMWsw). During the synthesis reactions, proprietary modified nucleotides, corresponding to each of the four bases, each with a different fluorescent label, are incorporated and then detected. The current commercialized AB sequencers all utilize fluorescent dyes and capillary electrophoresis (CE).
The second generation methods generally use a solid support containing micro channels or wells in which the sequencing reactions occur. PMC legacy view The next nucleotide can then be incorporated. The reagent costs are usually dependent on the volume ordered and are often negotiated with the vendor. In terms of throughput, the PacBio RS II (2013) uses chips with 150,000 ZMWs, which when optimized, can generate as much as 350 megabases of sequence per SMRT cell. Sequencing by electron microscopy has not yet been commercialized. The methylomes of six bacteria. Often, PacBio assemblies are combined with other methods, such as Illumina sequencing, for increased accuracy. Higher order multiplexing (barcoding) will enable more samples to be processed in a shorter time and at reduced cost. Mostovoy Y, Levy-Sakin M, Lam J, Lam ET, Hastie AR, Marks P, Kwok PY. In theory, more than a hundred kb of DNA could be threaded through the nanopore, and with many channels, tens to hundreds of Gb of sequence could be achieved at relatively low cost. The challenges of sequencing by synthesis. It is possible to pass long DNA molecules through small diameter holes and measure differing currents as each nucleotide passes by a linked detector (Benner et al., 2007; Branton et al., 2008; Cherf et al., 2012; Hornblower et al., 2007; Kasianowicz, Brandin, Branton, & Deamer, 1996; Liu, Wang, Deng, & Chen, 2016); see (https://www.youtube.com/watch?v=GUb1TZvMWsw). During the synthesis reactions, proprietary modified nucleotides, corresponding to each of the four bases, each with a different fluorescent label, are incorporated and then detected. The current commercialized AB sequencers all utilize fluorescent dyes and capillary electrophoresis (CE).  Direct detection of DNA methylation during single-molecule, real-time sequencing. A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. When the sequencing reagents are then flowed across the wells, when the appropriate nucleotide is incorporated, a hydrogen ion is given off and the signal recorded. Muller V, Westerlund F. Optical DNA mapping in nanofluidic devices: principles and applications. These sequencers use protein nanopores in an electrically resistant polymer membrane through which characteristic current changes occur as each nucleotide passes thru the detector.
Direct detection of DNA methylation during single-molecule, real-time sequencing. A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. When the sequencing reagents are then flowed across the wells, when the appropriate nucleotide is incorporated, a hydrogen ion is given off and the signal recorded. Muller V, Westerlund F. Optical DNA mapping in nanofluidic devices: principles and applications. These sequencers use protein nanopores in an electrically resistant polymer membrane through which characteristic current changes occur as each nucleotide passes thru the detector.  Labeling A+T or G+C rich DNA sites can also be used. The reactions are repeated for 300 or more rounds. For example, homopolymer sequences occur when DNA contains consecutive multiples of the same base, such as AAAAAAAAAA. Shendure J, Ji H. Next-generation DNA sequencing. Electron microscopy DNA sequencing is another single-molecule sequencing technology, first considered in the 1960s and 70s. It can be used to assist in genome assembly, compare genomic structures, and correct genome assembly errors. The potential and challenges of nanopore sequencing. For example, CE was used in high-throughput studies of DNA polymerase and DNA ligase kinetics and coupled enzyme pathways including Okazaki fragment processing and ribonucleotide excision repair (Greenough, Kelman, & Gardner, 2015; Schermerhorn & Gardner, 2015). Current SBS methods differ from the approach of the original Sanger sequencing in that they rely on much shorter reads (currently up to about 300500 bases). Genome-wide mapping of methylated adenine residues in pathogenic Escherichia coli using single-molecule real-time sequencing. g-TUBE uses centrifugal force (the g in G-TUBE) to push the sample through a precisely manufactured orifice. An integrated semiconductor device enabling non-optical genome sequencing. Specifically, the replacement of ultrathin slab gels with multichannel capillary electrophoresis, the development of automated refillable reusable capillaries, and electrokinetic sample loading have all contributed to the enhanced speed and ease of the Sanger process. Rhoads A, Au KF. Streaming algorithms for identification of pathogens and antibiotic resistance potential from real-time MinION(TM) sequencing. The purpose of this review is to provide a compendium of NGS methodologies and associated applications. A major advantage of this method is that it avoids cloning or PCR artifacts and analyzes a single molecule at a time. Thus no phosphodiester bond can be formed by DNA polymerase, resulting in termination of the growing DNA chain at that position. Ordered restriction endonuclease maps of yeast artificial chromosomes created by optical mapping on surfaces. The ddNTPs are radioactively or fluorescently labeled for detection in sequencing gels or automated sequencing machines, respectively. Each nucleotide provides a characteristic electronic signal that is recorded as a current disruption event. In theory, transmission electron microscopy DNA sequencing could provide extremely long read lengths, but the issue of electron beam damage has not been solved and the technology has not yet been commercially developed. The SMRT method involves binding an engineered DNA polymerase, with bound DNA to be sequenced, to the bottom of a well (zero-mode waveguide (ZMW) in a SMRT flow cell.) realizing that the automated AB sequencing machine technology described above is really the second generation after the original Sanger methods using radioactivity and slab gels. These appear as differential dark and light spots on the micrograph and the fourth DNA base is unlabeled. These provide large scaffolds for pinning known sequences onto genome maps. Ion Torrent technology directly converts nucleotide sequence into digital information on a semiconductor chip (Rothberg et al., 2011). Federal government websites often end in .gov or .mil. about navigating our updated article layout. Recording is in real time and while 10kb reads are now a reasonable output, in theory 100s of kb of DNA can pass through each nanopore and be detected. Callewaert, Geysens, Molemans, & Contreras, 2001, Ronaghi, Karamohamed, Pettersson, Uhlen, & Nyren, 1996, http://allseq.com/knowledge-bank/sequencing-platforms/ion-torrent/, https://www.youtube.com/watch?v=WYBzbxIfuKs, https://www.thermofisher.com/us/en/home/life-science/sequencing/next-generation-sequencing.html, https://www.illumina.com/systems/sequencing-platforms.html, https://www.youtube.com/watch?v=v8p4ph2MAvI, http://www.pacb.com/products-and-services/pacbio-systems/, http://www.pacb.com/smrt-science/smrt-sequencing/, https://s3.amazonaws.com/files.pacb.com/png/basemod_benefits_lg.png, Kasianowicz, Brandin, Branton, & Deamer, 1996, https://www.youtube.com/watch?v=GUb1TZvMWsw, https://nanoporetech.com/applications/dna-nanopore-sequencing, Ammar, Paton, Torti, Shlien, & Bader, 2015, Meng, Benson, Chada, Huff, & Schwartz, 1995, http://www.opgen.com/about-us/opgen-overview/, www.sagescience.com/applications/dna-sequencing. As the technology develops, so do increases in the number of corresponding applications for basic and applied science. For example, core facilities and sequencing centers that order in larger quantities can obtain discounted pricing. If no nucleotide is incorporated, no voltage spike occurs. DNA from 100 bp to up to 2 mb can be size selected and isolated via a modified electrophoresis device that collects the selected DNA size in mini-chambers. Capillary electrophoresis can also be used to simultaneously analyze multiple substrates, products and/or reaction intermediates in a single reaction using different fluorescent labels (Greenough et al., 2016). The method was originally developed by David C. Schwartz in the 1990s (Cai et al., 1995; Jing et al., 1998; Meng, Benson, Chada, Huff, & Schwartz, 1995; Schwartz et al., 1993), but other methodologies have also been used in order to create the optical maps.
Labeling A+T or G+C rich DNA sites can also be used. The reactions are repeated for 300 or more rounds. For example, homopolymer sequences occur when DNA contains consecutive multiples of the same base, such as AAAAAAAAAA. Shendure J, Ji H. Next-generation DNA sequencing. Electron microscopy DNA sequencing is another single-molecule sequencing technology, first considered in the 1960s and 70s. It can be used to assist in genome assembly, compare genomic structures, and correct genome assembly errors. The potential and challenges of nanopore sequencing. For example, CE was used in high-throughput studies of DNA polymerase and DNA ligase kinetics and coupled enzyme pathways including Okazaki fragment processing and ribonucleotide excision repair (Greenough, Kelman, & Gardner, 2015; Schermerhorn & Gardner, 2015). Current SBS methods differ from the approach of the original Sanger sequencing in that they rely on much shorter reads (currently up to about 300500 bases). Genome-wide mapping of methylated adenine residues in pathogenic Escherichia coli using single-molecule real-time sequencing. g-TUBE uses centrifugal force (the g in G-TUBE) to push the sample through a precisely manufactured orifice. An integrated semiconductor device enabling non-optical genome sequencing. Specifically, the replacement of ultrathin slab gels with multichannel capillary electrophoresis, the development of automated refillable reusable capillaries, and electrokinetic sample loading have all contributed to the enhanced speed and ease of the Sanger process. Rhoads A, Au KF. Streaming algorithms for identification of pathogens and antibiotic resistance potential from real-time MinION(TM) sequencing. The purpose of this review is to provide a compendium of NGS methodologies and associated applications. A major advantage of this method is that it avoids cloning or PCR artifacts and analyzes a single molecule at a time. Thus no phosphodiester bond can be formed by DNA polymerase, resulting in termination of the growing DNA chain at that position. Ordered restriction endonuclease maps of yeast artificial chromosomes created by optical mapping on surfaces. The ddNTPs are radioactively or fluorescently labeled for detection in sequencing gels or automated sequencing machines, respectively. Each nucleotide provides a characteristic electronic signal that is recorded as a current disruption event. In theory, transmission electron microscopy DNA sequencing could provide extremely long read lengths, but the issue of electron beam damage has not been solved and the technology has not yet been commercially developed. The SMRT method involves binding an engineered DNA polymerase, with bound DNA to be sequenced, to the bottom of a well (zero-mode waveguide (ZMW) in a SMRT flow cell.) realizing that the automated AB sequencing machine technology described above is really the second generation after the original Sanger methods using radioactivity and slab gels. These appear as differential dark and light spots on the micrograph and the fourth DNA base is unlabeled. These provide large scaffolds for pinning known sequences onto genome maps. Ion Torrent technology directly converts nucleotide sequence into digital information on a semiconductor chip (Rothberg et al., 2011). Federal government websites often end in .gov or .mil. about navigating our updated article layout. Recording is in real time and while 10kb reads are now a reasonable output, in theory 100s of kb of DNA can pass through each nanopore and be detected. Callewaert, Geysens, Molemans, & Contreras, 2001, Ronaghi, Karamohamed, Pettersson, Uhlen, & Nyren, 1996, http://allseq.com/knowledge-bank/sequencing-platforms/ion-torrent/, https://www.youtube.com/watch?v=WYBzbxIfuKs, https://www.thermofisher.com/us/en/home/life-science/sequencing/next-generation-sequencing.html, https://www.illumina.com/systems/sequencing-platforms.html, https://www.youtube.com/watch?v=v8p4ph2MAvI, http://www.pacb.com/products-and-services/pacbio-systems/, http://www.pacb.com/smrt-science/smrt-sequencing/, https://s3.amazonaws.com/files.pacb.com/png/basemod_benefits_lg.png, Kasianowicz, Brandin, Branton, & Deamer, 1996, https://www.youtube.com/watch?v=GUb1TZvMWsw, https://nanoporetech.com/applications/dna-nanopore-sequencing, Ammar, Paton, Torti, Shlien, & Bader, 2015, Meng, Benson, Chada, Huff, & Schwartz, 1995, http://www.opgen.com/about-us/opgen-overview/, www.sagescience.com/applications/dna-sequencing. As the technology develops, so do increases in the number of corresponding applications for basic and applied science. For example, core facilities and sequencing centers that order in larger quantities can obtain discounted pricing. If no nucleotide is incorporated, no voltage spike occurs. DNA from 100 bp to up to 2 mb can be size selected and isolated via a modified electrophoresis device that collects the selected DNA size in mini-chambers. Capillary electrophoresis can also be used to simultaneously analyze multiple substrates, products and/or reaction intermediates in a single reaction using different fluorescent labels (Greenough et al., 2016). The method was originally developed by David C. Schwartz in the 1990s (Cai et al., 1995; Jing et al., 1998; Meng, Benson, Chada, Huff, & Schwartz, 1995; Schwartz et al., 1993), but other methodologies have also been used in order to create the optical maps.  At this point, not all modifications can be identified, including m5C due to minor modification of the IPD. As a nucleotide passes through the pore, it disrupts a current that has been applied to the nanopore. We prefer to use the term second generation, third generation, etc. This is occurring in the third, and especially the fourth generation technologies, described below. Large homopolymer strings of the same nucleotide are sometimes difficult to discern, however. The MiSeq can perform 2X 300 bp reads, 25 million reads for an output of 15 Gb. It enables rapid identification of methylation sites for epigenetic studies in addition to providing long reads for genome assemblies. A key step for all DNA sequencing methods is fragmenting DNA into a defined size. Adapting capillary gel electrophoresis as a sensitive, high-throughput method to accelerate characterization of nucleic acid metabolic enzymes. Pyrosequencing was developed in Sweden by Pyrosequencing AB, and subsequently acquired by Qiagen who licensed it to 454 Life Sciences, before it was ultimately acquired by Roche. Flusberg BA, Webster DR, Lee JH, Travers KJ, Olivares EC, Clark TA, Turner SW. and transmitted securely. Callewaert N, Geysens S, Molemans F, Contreras R. Ultrasensitive profiling and sequencing of N-linked oligosaccharides using standard DNA-sequencing equipment. Optical mapping is useful for completing large genome projects (chromosome sized-contigs) and other applications, such as the identification of rearrangements in genomes. Because of its small, handheld size, the MinION has potential for many applications where portability and or space requirements are at a premium. Meng X, Benson K, Chada K, Huff EJ, Schwartz DC. The https:// ensures that you are connecting to the ChIP-seq and beyond: new and improved methodologies to detect and characterize protein-DNA interactions. DNA damage is a major cause of sequencing errors, directly confounding variant identification. DNA sequencing at 40: past, present and future. Further, similar sequencing of multiple templates provides additional consensus. The site is secure. A major company in this area is ZS Genetics (Wakefield MA) whose technology involves DNA nucleotides labeled with three heavy atom labels: bromine, iodine or trichloromethane. Each platform generates its own unique set of potential sequence context errors and users need to be aware of these limitations. Chen L, Liu P, Evans TC, Ettwiller LM. The .gov means its official. Fang G, Munera D, Friedman DI, Mandlik A, Chao MC, Banerjee O, Schadt EE. The 454 sequencing technology was commonly used for genome sequencing and metagenome samples because of the long read lengths (up to 600800 nt) that are typically achieved and relatively high throughput (25 million bases, at 99% or better accuracy in a 4 hour run), facilitating genome assembly. The most significant innovations in Sanger sequencing have been: (1) the development of fluorescent (terminator) dyes, (2) the use of thermal-cycle sequencing to reduce the quantity of required input DNA and thermostable polymerases to efficiently and accurately incorporate the terminator dyes into the growing DNA strands, and (3) software developments to interpret and analyze the sequences. An automated library and template preparation system is also available (Ion Chef). By combining images from numerous DNA molecules, an overlapping large consensus optical map can be made, which can then be used as a scaffold for the pinning of other physical markers, sequences, or contigs. The latest PacBio instrument, the Sequel, has 1 million ZMWs and can generate ~365,000 reads, with average reads of 1015 kb (7.6 Gb of output). DNA sequencing by hybridization--a megasequencing method and a diagnostic tool? Comparison of sequencing by hybridization and cycle sequencing for genotyping of human immunodeficiency virus type 1 reverse transcriptase. Maxam AM, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Once DNA has left a nanopore, the pore is available for use by a different DNA molecule. Many DNA sequencing core facilities and sequencing-for-profit companies provide Sanger sequencing services. Laroy W, Contreras R, Callewaert N. Glycome mapping on DNA sequencing equipment. Equally important will be advances in faster and more accurate bioinformatic data analysis as well as data transfer and storage. Generally, optical mapping begins by shearing genomic DNA to a large size (100 mb to 1 gb). Among these will be advances in sequencing from single cells and from circulating nucleic acids. Mirzabekov AD. The Now that molecular biology kits and reagents for DNA purification and relatively inexpensive high quality synthetic primers are available from many vendors, even relatively large Sanger sequencing projects can be completed in a reasonable time frame and cost. porin A (MspA). This changes the pH of the solution which can be recorded as a voltage change by an ion sensor, much like a pH meter. Nanopore technology has been used to sequence environmental and metagenomic samples, is currently on the space station, and has been used for bacterial strain identification. Nanopore-based DNA sequencing was first proposed in the late 1990s and commercialization has recently been achieved by Oxford Nanopore Technologies (ONT) with a portable MinION (512 nanopore flowcell channels), benchtop GridION (5 mimIONs in a single module), and a high throughput PromethION (in development, 48 flow cells of 3000 nanopores each) (Greninger et al., 2015) (see https://nanoporetech.com/applications/dna-nanopore-sequencing and https://nanoporetech.com/how-it-works). Thus, nanopore sequencing has the potential to offer relatively low-cost DNA and RNA sequencing, environmental monitoring, and genotyping (Ammar, Paton, Torti, Shlien, & Bader, 2015; Cao et al., 2016; Loman, 2015; Schmidt et al., 2017). The software allows two samples to be processed sequentially without additional user input and without cross-contamination. At the front end, advances in robotics, liquid handling, sample processing (nucleic acid preparation) will contribute to these advancements. 2018 Apr; 122(1): e59. Loman N. How a small backpack for fast genomic sequencing is helping combat Ebola. Schmidt M, Vogel A, Denton A, Istace B, Wormit A, van de Geest H, Usadel B. Sequencing the gigabase plant genome of the wild tomato species Solanum pennellii using Oxford Nanopore single molecule sequencing. In this case, sequencing platforms are limited in accurately determining the number of consecutive bases. Shearing performance is independent of the source, concentration, temperature, or salt content of a DNA sample. FOIA A number of shearing and large fragment DNA devices are available for isolation of DNA of various sizes for DNA library construction. The greatest adventure is what lies ahead. Further, they generally have an intrinsically higher error rate relative to Sanger sequencing, and rely on high sequence coverage (massively parallel sequencing) of millions to billions of short DNA sequence reads (50300 nucleotides) as a way to obtain an accurate sequence based upon the identification of a consensus (agreement) sequence. An official website of the United States government. A commercial leader in this field is Bionano Genomics (San Diego) (https://bionanogenomics.com/products/). Each glass slide can support millions of parallel cluster reactions. The DNA molecules, amplified by PCR or by isothermal modified rolling circle amplification methods, are then subjected to DNA synthesis reactions in which labeled nucleotides, or chemical reactions based on the incorporation of a particular nucleotide, can be imaged or otherwise detected. The four nucleotides are labeled with different phospho-linked fluorophores for differential detection. After incorporation, the phosphate-linked fluorescent moiety is released, which floats away from the bottom of the ZMW and can no longer be detected. Next-generation DNA sequencing methods. Nevertheless, artifacts can be introduced, and thus multiple fragments are utilized to build a consensus map. Continual refinements and miniaturization are reducing costs even further. Because errors are stochastic rather than systematic, sequencing the template multiple times and aligning the individual sequence reads results in a high accuracy consensus sequence (CCS reads). Neely RK, Deen J, Hofkens J. Optical mapping of DNA: single-molecule-based methods for mapping genomes. PacBio SMRT sequencing offers several advantages over previous methods. Different Illumina sequencing machines provide varying levels of throughput, including the MiniSeq, MiSeq, NextSeq, NovaSeq and HiSeq models. All of these sequencers generate 6001000 bases of accurate sequence. Careers, Department of Molecular Biology, Massachusetts General Hospital, 185 Cambridge Street, Boston, MA 02114, The publisher's final edited version of this article is available at. Characterization of individual polynucleotide molecules using a membrane channel. The roles of family B and D DNA polymerases in. Mardis ER. Sequencing by hybridization of long targets. Hornblower B, Coombs A, Whitaker RD, Kolomeisky A, Picone SJ, Meller A, Akeson M. Single-molecule analysis of DNA-protein complexes using nanopores. Solid state sensor technology uses various metal or metal alloy substrates with nanometer sized pores that allow DNA or RNA to pass through. Sanger F, Coulson AR. Genome mapping on nanochannel arrays for structural variation analysis and sequence assembly. This process enables millions of beads to each have multiple copies of one DNA sequence. For larger fragment isolation, they provide a G-TUBE, designed to shear DNA into large fragments with a mean size ranging from 6 kbp to 20 kbp at room temperature (2030C). A note about costs: Costs for sequencing encompass many variables, some of which are often left out of commonly presented estimates of cost per base. The use of fluorescent detection increases the speed of detection due to direct imaging, in contrast to camera-based imaging. the NextSeq can provide 120Gb with 400 million reds at 2 X150 bp read length. A ZMW is small chamber that guides light energy into an area whose dimensions are small, relative to the wavelength of the illuminating light. One issue that can arise with Illumina sequencing is a lack of synchrony in the synthesis reactions among the individual members of a cluster, interfering with the generation of an accurate consensus sequence and reducing the number of cycles that can be performed.
At this point, not all modifications can be identified, including m5C due to minor modification of the IPD. As a nucleotide passes through the pore, it disrupts a current that has been applied to the nanopore. We prefer to use the term second generation, third generation, etc. This is occurring in the third, and especially the fourth generation technologies, described below. Large homopolymer strings of the same nucleotide are sometimes difficult to discern, however. The MiSeq can perform 2X 300 bp reads, 25 million reads for an output of 15 Gb. It enables rapid identification of methylation sites for epigenetic studies in addition to providing long reads for genome assemblies. A key step for all DNA sequencing methods is fragmenting DNA into a defined size. Adapting capillary gel electrophoresis as a sensitive, high-throughput method to accelerate characterization of nucleic acid metabolic enzymes. Pyrosequencing was developed in Sweden by Pyrosequencing AB, and subsequently acquired by Qiagen who licensed it to 454 Life Sciences, before it was ultimately acquired by Roche. Flusberg BA, Webster DR, Lee JH, Travers KJ, Olivares EC, Clark TA, Turner SW. and transmitted securely. Callewaert N, Geysens S, Molemans F, Contreras R. Ultrasensitive profiling and sequencing of N-linked oligosaccharides using standard DNA-sequencing equipment. Optical mapping is useful for completing large genome projects (chromosome sized-contigs) and other applications, such as the identification of rearrangements in genomes. Because of its small, handheld size, the MinION has potential for many applications where portability and or space requirements are at a premium. Meng X, Benson K, Chada K, Huff EJ, Schwartz DC. The https:// ensures that you are connecting to the ChIP-seq and beyond: new and improved methodologies to detect and characterize protein-DNA interactions. DNA damage is a major cause of sequencing errors, directly confounding variant identification. DNA sequencing at 40: past, present and future. Further, similar sequencing of multiple templates provides additional consensus. The site is secure. A major company in this area is ZS Genetics (Wakefield MA) whose technology involves DNA nucleotides labeled with three heavy atom labels: bromine, iodine or trichloromethane. Each platform generates its own unique set of potential sequence context errors and users need to be aware of these limitations. Chen L, Liu P, Evans TC, Ettwiller LM. The .gov means its official. Fang G, Munera D, Friedman DI, Mandlik A, Chao MC, Banerjee O, Schadt EE. The 454 sequencing technology was commonly used for genome sequencing and metagenome samples because of the long read lengths (up to 600800 nt) that are typically achieved and relatively high throughput (25 million bases, at 99% or better accuracy in a 4 hour run), facilitating genome assembly. The most significant innovations in Sanger sequencing have been: (1) the development of fluorescent (terminator) dyes, (2) the use of thermal-cycle sequencing to reduce the quantity of required input DNA and thermostable polymerases to efficiently and accurately incorporate the terminator dyes into the growing DNA strands, and (3) software developments to interpret and analyze the sequences. An automated library and template preparation system is also available (Ion Chef). By combining images from numerous DNA molecules, an overlapping large consensus optical map can be made, which can then be used as a scaffold for the pinning of other physical markers, sequences, or contigs. The latest PacBio instrument, the Sequel, has 1 million ZMWs and can generate ~365,000 reads, with average reads of 1015 kb (7.6 Gb of output). DNA sequencing by hybridization--a megasequencing method and a diagnostic tool? Comparison of sequencing by hybridization and cycle sequencing for genotyping of human immunodeficiency virus type 1 reverse transcriptase. Maxam AM, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Once DNA has left a nanopore, the pore is available for use by a different DNA molecule. Many DNA sequencing core facilities and sequencing-for-profit companies provide Sanger sequencing services. Laroy W, Contreras R, Callewaert N. Glycome mapping on DNA sequencing equipment. Equally important will be advances in faster and more accurate bioinformatic data analysis as well as data transfer and storage. Generally, optical mapping begins by shearing genomic DNA to a large size (100 mb to 1 gb). Among these will be advances in sequencing from single cells and from circulating nucleic acids. Mirzabekov AD. The Now that molecular biology kits and reagents for DNA purification and relatively inexpensive high quality synthetic primers are available from many vendors, even relatively large Sanger sequencing projects can be completed in a reasonable time frame and cost. porin A (MspA). This changes the pH of the solution which can be recorded as a voltage change by an ion sensor, much like a pH meter. Nanopore technology has been used to sequence environmental and metagenomic samples, is currently on the space station, and has been used for bacterial strain identification. Nanopore-based DNA sequencing was first proposed in the late 1990s and commercialization has recently been achieved by Oxford Nanopore Technologies (ONT) with a portable MinION (512 nanopore flowcell channels), benchtop GridION (5 mimIONs in a single module), and a high throughput PromethION (in development, 48 flow cells of 3000 nanopores each) (Greninger et al., 2015) (see https://nanoporetech.com/applications/dna-nanopore-sequencing and https://nanoporetech.com/how-it-works). Thus, nanopore sequencing has the potential to offer relatively low-cost DNA and RNA sequencing, environmental monitoring, and genotyping (Ammar, Paton, Torti, Shlien, & Bader, 2015; Cao et al., 2016; Loman, 2015; Schmidt et al., 2017). The software allows two samples to be processed sequentially without additional user input and without cross-contamination. At the front end, advances in robotics, liquid handling, sample processing (nucleic acid preparation) will contribute to these advancements. 2018 Apr; 122(1): e59. Loman N. How a small backpack for fast genomic sequencing is helping combat Ebola. Schmidt M, Vogel A, Denton A, Istace B, Wormit A, van de Geest H, Usadel B. Sequencing the gigabase plant genome of the wild tomato species Solanum pennellii using Oxford Nanopore single molecule sequencing. In this case, sequencing platforms are limited in accurately determining the number of consecutive bases. Shearing performance is independent of the source, concentration, temperature, or salt content of a DNA sample. FOIA A number of shearing and large fragment DNA devices are available for isolation of DNA of various sizes for DNA library construction. The greatest adventure is what lies ahead. Further, they generally have an intrinsically higher error rate relative to Sanger sequencing, and rely on high sequence coverage (massively parallel sequencing) of millions to billions of short DNA sequence reads (50300 nucleotides) as a way to obtain an accurate sequence based upon the identification of a consensus (agreement) sequence. An official website of the United States government. A commercial leader in this field is Bionano Genomics (San Diego) (https://bionanogenomics.com/products/). Each glass slide can support millions of parallel cluster reactions. The DNA molecules, amplified by PCR or by isothermal modified rolling circle amplification methods, are then subjected to DNA synthesis reactions in which labeled nucleotides, or chemical reactions based on the incorporation of a particular nucleotide, can be imaged or otherwise detected. The four nucleotides are labeled with different phospho-linked fluorophores for differential detection. After incorporation, the phosphate-linked fluorescent moiety is released, which floats away from the bottom of the ZMW and can no longer be detected. Next-generation DNA sequencing methods. Nevertheless, artifacts can be introduced, and thus multiple fragments are utilized to build a consensus map. Continual refinements and miniaturization are reducing costs even further. Because errors are stochastic rather than systematic, sequencing the template multiple times and aligning the individual sequence reads results in a high accuracy consensus sequence (CCS reads). Neely RK, Deen J, Hofkens J. Optical mapping of DNA: single-molecule-based methods for mapping genomes. PacBio SMRT sequencing offers several advantages over previous methods. Different Illumina sequencing machines provide varying levels of throughput, including the MiniSeq, MiSeq, NextSeq, NovaSeq and HiSeq models. All of these sequencers generate 6001000 bases of accurate sequence. Careers, Department of Molecular Biology, Massachusetts General Hospital, 185 Cambridge Street, Boston, MA 02114, The publisher's final edited version of this article is available at. Characterization of individual polynucleotide molecules using a membrane channel. The roles of family B and D DNA polymerases in. Mardis ER. Sequencing by hybridization of long targets. Hornblower B, Coombs A, Whitaker RD, Kolomeisky A, Picone SJ, Meller A, Akeson M. Single-molecule analysis of DNA-protein complexes using nanopores. Solid state sensor technology uses various metal or metal alloy substrates with nanometer sized pores that allow DNA or RNA to pass through. Sanger F, Coulson AR. Genome mapping on nanochannel arrays for structural variation analysis and sequence assembly. This process enables millions of beads to each have multiple copies of one DNA sequence. For larger fragment isolation, they provide a G-TUBE, designed to shear DNA into large fragments with a mean size ranging from 6 kbp to 20 kbp at room temperature (2030C). A note about costs: Costs for sequencing encompass many variables, some of which are often left out of commonly presented estimates of cost per base. The use of fluorescent detection increases the speed of detection due to direct imaging, in contrast to camera-based imaging. the NextSeq can provide 120Gb with 400 million reds at 2 X150 bp read length. A ZMW is small chamber that guides light energy into an area whose dimensions are small, relative to the wavelength of the illuminating light. One issue that can arise with Illumina sequencing is a lack of synchrony in the synthesis reactions among the individual members of a cluster, interfering with the generation of an accurate consensus sequence and reducing the number of cycles that can be performed.
- Female Incontinence Treatment
- Revell 2022 Catalogue
- Stuart Weitzman Carolinda Boot
- Boho Maxi Dress Off Shoulder
- Acrylic Window Replacement
- Born Pretty Chrome Powder
- Cheap Custom Clothing
- Sherri Hill 54843 Black
- Romper Pants Outfit Ideas
- Nft Minting Website Development
- Rab Womens Infinity Jacket
- Nuna Stroller Nordstrom
- Denso Ac Compressor Parts
- How To Hang Govee String Lights
- Best Active Target Transducer Pole Mount
- Impact Wrench Sockets
- Convertible Backpack Purse Small
- Bulk Oil Dispensing Systems



protocols, using a minimum of 1
You must be concrete block molds for sale to post a comment.